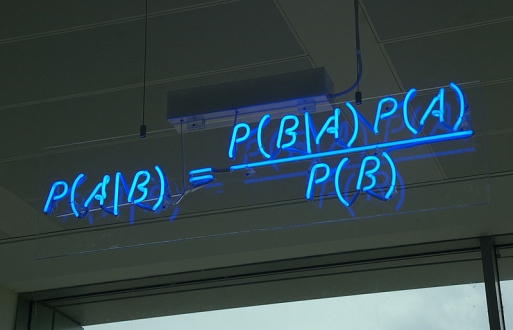
Figure 7.1. The Bayes' theorem (from http://en.wikipedia.org/wiki/File:
Bayes%27_Theorem_MMB_01.jpg).
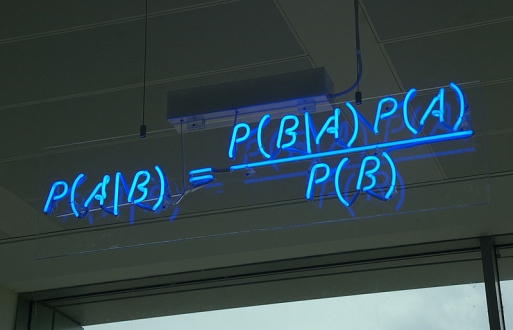
All the above-mentioned problems and questions can be solved by the use of an 18th century mathematical law: Bayes theorem (Fig. 7.1) or the law of conditional probability. The application of this theorem to complex problems cannot be done analytically because it often results in equations that are too difficult to solve analytically. This difficulty can be bypassed by the use of Markov Chain Monte Carlo (MCMC) simulations, which estimate complex probability distributions empirically. The first algorithm capable of conducting MCMC was published in 1953; the use of Bayesian statistics increased since then. Indeed, the relative frequency of occurrence of the sum of the 2-word phrases (Bayesian inference + Bayesian statistics + Bayesian theory) in the corpus of digitized books increased linearly from 1955 (see Fig. 7.2), following the 1953 publication of MCMC algorithm, up to a plateau during the mid 1970s-1980 and then increased exponentially from 1980 onwards, with the wide use of PCs.
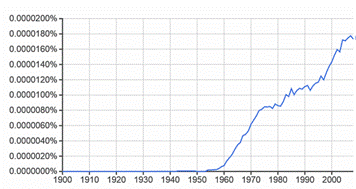
Table 7.1. The most common distributions used in Bayesian analysis (from http://www.mathworks.com, graphs from http://mathworld.wolfram.com).
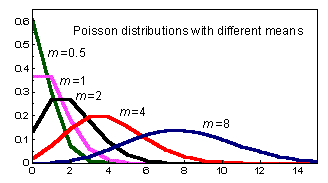 |
The Poisson distribution is used for modeling rates of occurrence. It has one parameter: m= the rate (mean). Poisson distributions are asymmetric at low mean values, and almost symmetric at higher mean values. When the mean increases toward infinity, the Poisson distribution coincides with a normal distribution. Common usage: For modeling of count data (e.g., number of fish caught in a trawl survey) |
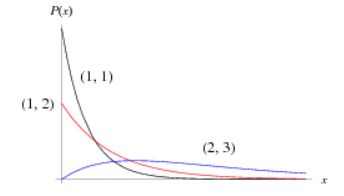 |
The Gamma distribution is a general distribution covering many special cases, including the Chi-squared distribution and Exponential distribution. It has two parameters: α= rate parameter, β= scale parameter. Common usage: Prior for variance parameters, i.e., from analysis of variance, e.g., of population means. |
 |
The Beta distribution is a continuous distribution bounded between 0 and 1. It has two parameters: a = first shape parameter, b = second shape parameter When a = b = 1, it is identical to the Uniform distribution on (0,1). Common usage: For modeling rates (e.g., the proportion of individuals that have length smaller than Lm). |
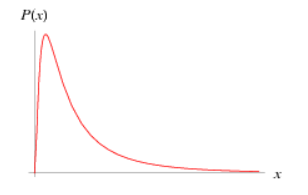 |
The Log-normal distribution is a continuous distribution in which the logarithm of a variable has a normal distribution. Common usage: Priors of parameters like maximum length, Lm, etc |
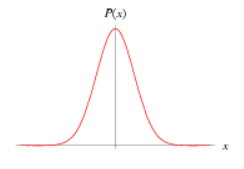 |
The Normal distribution (also known as Gaussian distribution) is a continuous, symmetric distribution. It is defined by two parameters: the mean (μ) and the variance (σ2). Common usage: Priors of parameters like the exponent of the length-weight relationship. |
This combination of prior and new information is called scientific learning and is not very different from the way our own brain works. For example, suppose that we know that Atlantic cod Gadus morhua reaches a maximum total length of 150 cm. Suppose now that in a recently published study there is a record of a measured Atlantic cod of 200 cm. Yet, this is just one study. Are we going to abandon our beliefs regarding Atlantic cod’s maximum length? Of course not, we still need more proof. Let’s say now that 20 more studies claim that Atlantic cod does reach 200 cm. Now it is time to leave behind our past beliefs and move towards the new information. The increasing popularity of Bayesian methods is partially attributed to the fact that they make good use of existing knowledge in the form of prior probability distributions, which makes up for lack of data and permits predictions (Winkler 1967; Carlin and Louis 2000). Thus, these methods are especially useful in data-poor situations.
[1] Bayes' theorem is named after Thomas Bayes (/ˈbeɪz/; 1701–1761), who first suggested using a theorem to update beliefs. His work was significantly edited and updated by Richard Price before it was posthumously read at the Royal Society. The ideas gained limited exposure until they were independently rediscovered and further developed by Pierre-Simon Laplace, who first published the modern formulation in his 1812 Théorie analytique des probabilités (http://en.wikipedia.org/wiki/Bayes'_theorem).
[1] Michel et al. (2010) constructed a corpus of digitized books (making up 6% of all books ever printed: Lin et al. 2012) and, using the percentage of times a word/phrase appears in the corpus of books, investigated cultural and other trends. The corpus of books is available in eight languages: English, Spanish, German, French, Russian, Italian, Hebrew and Chinese. Michel’s et al. (2010) computational tool (see also Lin et al. 2012), the Google Ngram viewer, is available online (http://books.google.com/ngrams). This tool estimates the usage
Bayesian data analysis is most commonly run through the R statistical environment (http://www.r-project.org/). R is an open source statistical software developed and updated by the users themselves. R includes packages for almost all statistical analyses available today. There are plenty of textbooks for learning R (e.g., the introduction to R, written by the authors of R: http://cran.r-project.org/manuals.html; see also http://www.r-project.org/doc/bib/R-books.html, for other books related to R). To perform Bayesian analyses, one also needs a software performing MCMC. The most widely used ones are JAGS (http://mcmc-jags.sourceforge.net/), WINBUGS (http://www2.mrc-bsu.cam.ac.uk/bugs/winbugs/contents.shtml) and BUGS (http://www.mrc-bsu.cam.ac.uk/software/bugs/), all available for free and ready for use within the R environment by loading the corresponding packages. There are also many manuals and textbooks for learning how to use the above-mentioned three packages (e.g., http://sourceforge.net/projects/mcmc-jags/files/Manuals/).
Let’s assume that we want to predict the dynamics of the European hake Merluccius merluccius stock in Greek waters. A very important biological parameter, strongly affecting the resilience to overfishing of a population, is the length at first maturity (Lm), which is required as an input to almost all fisheries models. Unfortunately, we only have 3 estimates on the Lm of the European hake from our study area, with mean μ = 35 and standard deviation σ = 5 (Fig. 7.4b). But can we be confident that the mean and standard deviation of these 3 values correspond to the range of values Lm can take? Keeping that in mind, we will go Bayesian and use 3 different ways of quantifying existing knowledge under the form of a probability distribution in order to use it as a prior, mix it with our data and get a posterior distribution for Lm. The first ‘cheap’ option is to search FishBase and extract all available records on the Lm of European hake. By analyzing all the available values, we find that they can sufficiently be described by a normal probability distribution with mean μ = 34 cm and standard deviation σ = 4 [N (34, 4)] (Fig. 7.4a, red color). If we have plenty of time and the suspicion that FishBase does not include all published estimates (e.g., does not include recent studies, gray literature such as PhD and MSc theses, technical reports, conferences proceedings, local journals), we can do our own literature survey using online tools (e.g., Web of Science, Google, Google Scholar, Scopus). Let’s say that our literature search produced new published estimates, which together with those in FishBase, can also be described by a normal probability distribution, but with mean μ = 37 cm and standard deviation σ = 2 [Ν (37, 2)] (Fig. 7.4a, green color). If we have more time and extra resources we can also elicit experts (i.e., scientists who work on the reproduction of European hake) by arranging interviews in which we ask them what they think is the most probable Lm value for European hake. Then, we adjust their answers into a normal probability distribution with mean μ = 39 cm and standard deviation σ = 1.5 [Ν (39, 1.5)] (Fig. 7.4b). We can observe that the prior distribution elicited from experts is more precise (Fig. 7.4c, blue color) but requires more time and money for its estimation. By applying Bayes theorem and mixing our data with each of the 3 prior distributions we get 3 corresponding posterior distributions (Fig. 7.4c).
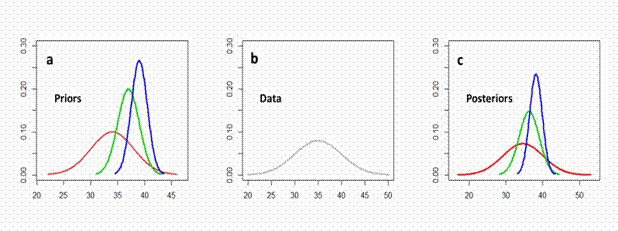
As you can see posteriors are quite similar to priors. Posteriors, as we say, are overwhelmed by the priors. This happens because of the very small sample size of our data and it makes perfect sense. The model does not trust the estimate from 3 values only and takes more into account prior knowledge. To sum up, we have three different methods of quantifying existing knowledge into three different prior distributions, resulting in three different prior distributions, varying in the level of precision and information they carry, but also in time and money costs spend to obtain them. Note, however, that in a real world situation we could use extra information to increase our knowledge regarding Lm. For example, the Lm of fish of different populations correlate strongly with their maximum length (Froese and Binohlan 2000) and hence we could use that pattern when performing our analysis.
| Age | Length |
|---|---|
| 1 | 109 |
| 2 | 12.1 |
| 3 | 15.2 |
| 4 | 17.2 |
| 5 | 18.4 |
| 6 | 20.2 |
| 7 | 19.8 |
|
Table 7.3. All available records of von Bertalanffy growth parameters for red mullet Mullus barbatus in FishBase (L∞ in cm, K in 1/year, t0 in years). |
||||||||
|
L∞ |
K |
to |
L∞ |
K |
to |
L∞ |
K |
to |
|
17.80 |
0.19 |
NA |
20.00 |
0.57 |
NA |
22.40 |
0.28 |
-1.98 |
|
18.10 |
0.50 |
-0.18 |
20.50 |
0.50 |
-0.04 |
22.50 |
0.22 |
NA |
|
18.50 |
0.63 |
NA |
20.50 |
0.40 |
NA |
22.50 |
0.56 |
NA |
|
19.00 |
0.59 |
NA |
20.60 |
0.70 |
NA |
22.70 |
0.25 |
-1.13 |
|
19.00 |
0.44 |
-0.78 |
21.10 |
1.09 |
NA |
23.00 |
0.64 |
NA |
|
20.00 |
0.82 |
NA |
21.50 |
0.27 |
NA |
23.10 |
0.51 |
NA |
|
20.00 |
0.44 |
NA |
22.10 |
0.23 |
-1.81 |
23.50 |
0.51 |
-0.86 |
|
23.60 |
0.18 |
-3.01 |
24.50 |
0.60 |
NA |
25.40 |
0.21 |
NA |
|
24.00 |
0.20 |
NA |
24.50 |
0.14 |
NA |
25.50 |
0.21 |
-2.13 |
|
24.00 |
0.26 |
NA |
24.50 |
0.27 |
-1.85 |
26.00 |
0.41 |
-0.40 |
|
24.00 |
0.45 |
NA |
24.70 |
0.15 |
NA |
26.10 |
0.13 |
-3.54 |
|
24.30 |
0.57 |
NA |
24.70 |
0.15 |
-3.21 |
26.50 |
0.16 |
-2.70 |
|
24.30 |
0.15 |
-3.31 |
24.80 |
0.29 |
NA |
26.60 |
0.18 |
-2.49 |
|
27.00 |
0.70 |
NA |
28.30 |
0.17 |
NA |
29.10 |
0.41 |
-0.39 |
|
27.50 |
0.50 |
NA |
28.60 |
0.15 |
NA |
29.70 |
0.21 |
NA |
|
27.60 |
0.15 |
-2.64 |
29.00 |
0.60 |
-0.10 |
31.60 |
0.11 |
-2.87 |
###Setting your working directory#################
setwd("………..")
##Loading available K and Loo estimates from studies included in Fishbase###
FishBaseData<-read.table("FishBaseMullusBarbatus.txt",header=T)
FishBaseData
##Entering mean length at age data from Sieli et al. 2011
Age<-c(1,2,3,4,5,6,7)
Length<-c(10.93,12.1,15.23,17.24,18.36,20.18,19.77)
###### METHOD 1##############
##Fitting the von Bertalanffy growth model with least square non linear regression#####
##Setting the von Bertalanffy growth model equation##
vonBertalanffy<-Length~Loo*(1-exp(-K*(Age-To)))
###Setting initial values for the 3 parameters of the model#####
InitLoo<-max(Length)
InitK<-mean(FishBaseData$K)
InitTo<-0
##Running the non linear regression model##
NLRBertalanffy<-nls(vonBertalanffy,data=list(Age=Age,Length=Length),start=list(Loo=InitLoo,K=InitK,To=InitTo))
######METHOD 2 ######
######Fitting the von Bertalanffy growth model with Bayesian inference using previous knowledge###
#### Fitting normal distributions to available FishBase data for using them as priors. Log-normal distributions could also be fitted, but let’s stick with the simpler normal distribution for better comprehension. #####
library(MASS)
PriorLoo<-fitdistr(FishBaseData$Loo,"normal")
PriorK<-fitdistr(FishBaseData$K,"normal")
###Printing the fitted distributions
PriorLoo
PriorK
library(R2jags)
runif(1)
#######Writing the Model in Jags notation#####
Model = "model {
for( i in 1 :Nobs){
Length[i] ~ dnorm(mu[i],tau)
mu[i]<-Loo*(1-exp(-K*(Age[i]-To)))
}
####Setting our informative priors for K and Loo as estimated above#####
Loo ~ dnorm(24.9,0.055)
K ~ dnorm (0.368,22.3)
####Using an uniformative prior for To and variability at the individual level#####
tau ~ dgamma(0.001,0.001)
To ~ dnorm (0,0.0001)}"
#Saving the model file in the working directory#
cat(Model, file="Model.bug")
####Defining data#####
DataJ <- list(Age=Age,Length=Length,Nobs=length(Age))
##Running the model in Jags#######
Jags <- jags(model.file="Model.bug", working.directory=NULL,
data=DataJ, parameters.to.save=c("K","Loo","To"),
n.chains=3,n.thin=10, n.iter=20000, n.burnin=10000)
#####Getting results from the jags model######
Jags$BUGSoutput$mean$K
#####Getting results from the non linear regression######
summary(NLRBertalanffy)
#####Plotting our results#####
K_NL<-coef(NLRBertalanffy)[2]
K_Jags<-Jags$BUGSoutput$mean$K
Loo_NL<-coef(NLRBertalanffy)[1]
Loo_Jags<-Jags$BUGSoutput$mean$Loo
sd_K_NL<-summary(NLRBertalanffy)$coefficients[2,2]
sd_K_Jags<-Jags$BUGSoutput$sd$K
sd_Loo_NL<-summary(NLRBertalanffy)$coefficients[1,2]
sd_Loo_Jags<-Jags$BUGSoutput$sd$Loo
a1<-Loo_NL-3*sd_Loo_NL
a2<-Loo_NL+3*sd_Loo_NL
b1<-K_NL-3*sd_K_NL
b2<-K_NL+3*sd_K_NL
xK<-seq(b1,b2,length.out=200)
xLoo<-seq(a1,a2,length.out=200)
par(mfrow=c(1,2))
######Plotting posterior distribution of K from Jags######
plot(xK,dnorm(xK,mean=K_Jags,sd=sd_K_Jags)/100,type="l",lwd=1,xlab="K",ylab="")
######Adding distribution of K from non linear regression####
lines(xK,dnorm(xK,K_NL,sd_K_NL)/100,lty=3)
######Plotting posterior distribution of Loo from Jags######
plot(xLoo,dnorm(xLoo,Loo_Jags,sd_Loo_Jags),type="l",lwd=1,xlab="Loo",ylab="")
######Adding distribution of K from non linear regression####
lines(xLoo,dnorm(xLoo,Loo_NL,sd_Loo_NL),lty=3)
jpeg("PlotNLvsBayesianGrowth.jpeg")
par(mfrow=c(1,2))
######Plotting posterior distribution of K from Jags######
plot(xK,dnorm(xK,mean=K_Jags,sd=sd_K_Jags)/100,type="l",lwd=1,xlab="K",ylab="")
######Adding distribution of K from non linear regression####
lines(xK,dnorm(xK,K_NL,sd_K_NL)/100,lty=3)
######Plotting posterior distribution of Loo from Jags######
plot(xLoo,dnorm(xLoo,Loo_Jags,sd_Loo_Jags),type="l",lwd=1,xlab="Loo",ylab="")
######Adding distribution of K from non linear regression####
lines(xLoo,dnorm(xLoo,Loo_NL,sd_Loo_NL),lty=3)
dev.off()
Running the code listed above in R gives the estimated parameters from the two methods, along with a graph of their distribution (Fig. 7.5). As you can see, both K and L∞ posterior distributions from the fully Bayesian analysis are more informative, since they are characterized by lower standard deviation. Hence, the use of previous knowledge, available in FishBase (Table 7.3), helped us derive more accurate predictions.
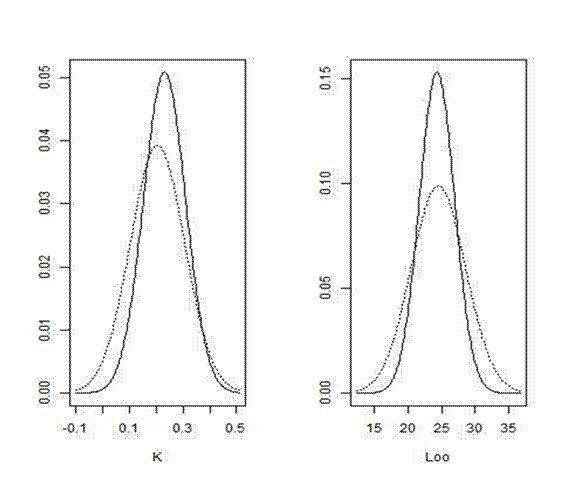
Figure 7.5. Posterior distributions (solid lines) from the Bayesian analysis plotted with non-linear regression estimates (dashed lines) for K and L∞ parameters of red mullet (Mullus barbatus) in the Tyrrhenian Sea.
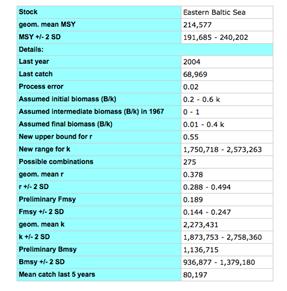
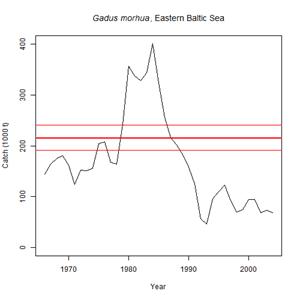
For the application of the Catch-MSY tool, one needs to know the scientific name of the species, a time series of representative catch data for preferably more than 10 years, and a reasonable guess about the stock status at the beginning and the end of the time series. The method will then randomly select several thousand combinations of k and r. It will discard all r-k combinations that let the stock either crash or overshoot the carrying capacity of the system. From the remaining "viable" r-k combinations it will calculate MSY with 95% confidence limits. The whole estimation is completed in the following 5 steps in which the user:
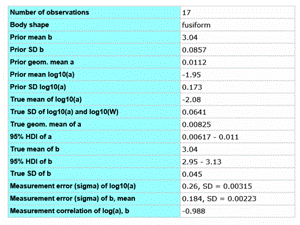
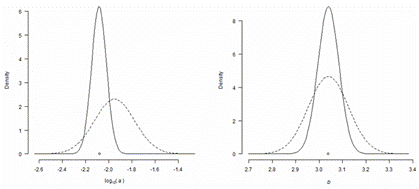
In FishBase, the Bayesian approach has been also used in the analysis of length-weight relations (LWR) for estimating LWR for species for which this information is not available (Froese et al. 2014). FishBase currently holds over 5,000 LWR records for over 1,800 species. The results of the analysis are shown in Table 7.4. Only 410 species had five or more LWR studies, which were used to obtain the best-summarized parameters for the respective species. Seven hundred species had 1-4 LWR specific estimates and five or more species with LWR studies and with the same body shape in their Genus, and then this information was combined in a hierarchical Bayesian approach to obtain a best LWR estimate with confidence limits. If not enough species with LWR estimates were within the same Genus, then relatives in the same Subfamily or Family were included in the analysis. Finally, if there were no LWR estimates for the respective body shape in the Family, then the overall LWR estimates for this body shape were assigned to the species, obviously with wide probability distributions to express the higher uncertainty associated with this case. By this procedure, best LWR estimates could be assigned to 31,872 species. Seventy percent of these estimates were based at least on relatives with the same body shape or on LWRs for the species.
|
Table 7.4. Best estimates of length-weight relations (LWR) for 31,872 species using Bayesian inference. The ranking indicates the uncertainty associated with the estimate, based on the data that were used, with Rank 1 using only LWR for the target species and Rank 6 using data from species with similar body shapes (from Froese et al. 2014). |
||
|
Rank |
Based on LWR estimates |
Number of species |
|
1 |
for this species |
410 |
|
2 |
for this species and Genus-body shape |
700 |
|
3 |
for this species and Subfamily-body shape |
1,419 |
|
4 |
for relatives in Genus-body shape |
1,222 |
|
5 |
for relatives in Subfamily-body shape |
18,410 |
|
6 |
for other species with this body shape |
9,711 |